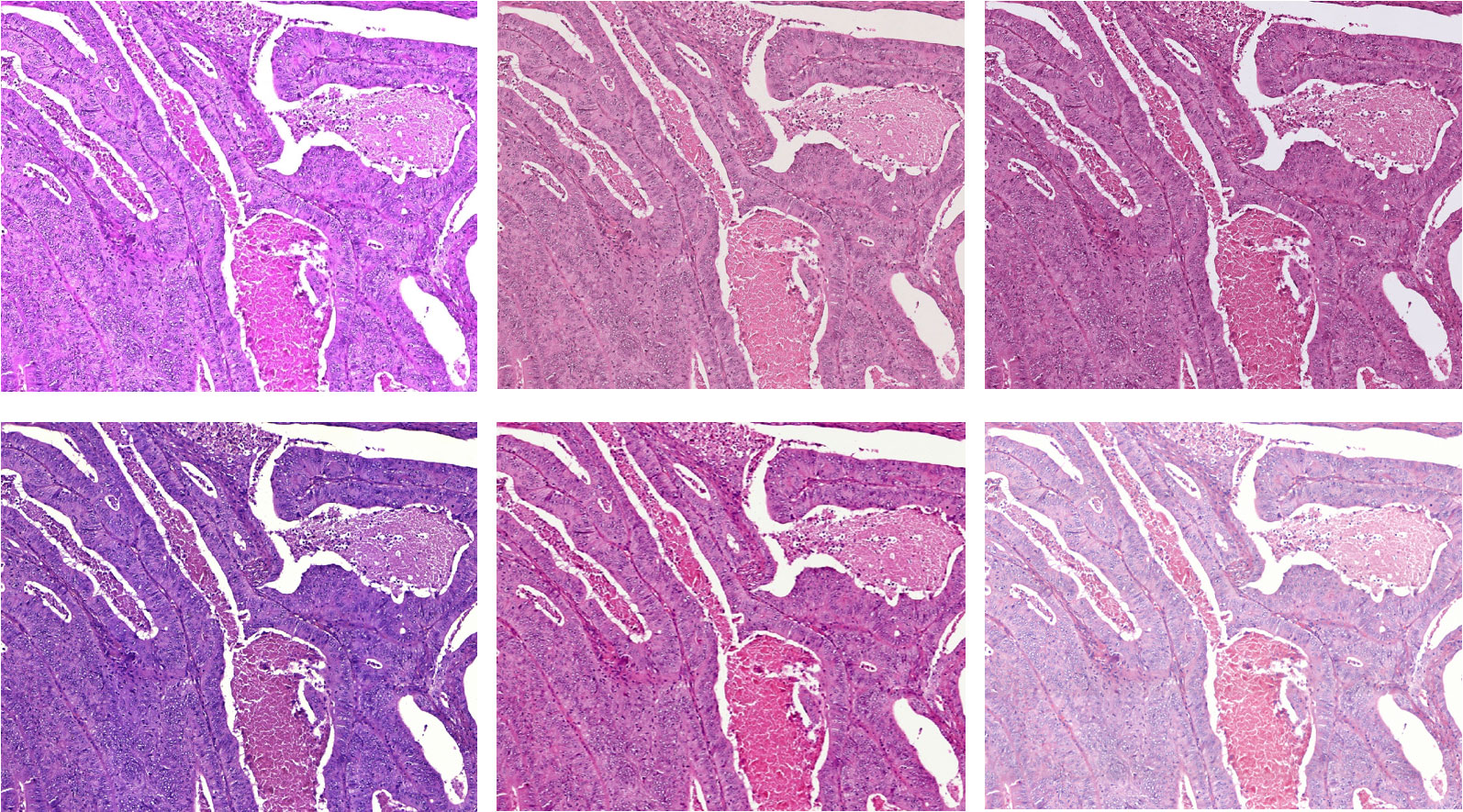
A significant challenge in the development of AI-based diagnostic support lies in the strong heterogeneity of digital tissue sections, which results, for example, from differences in sample preparation between different clinics or from the use of tissue scanners from different manufacturers. As a result, the images sometimes vary greatly, for example in color tone, saturation or resolution.
It would also be preferable for the procedures to be easily adaptable to new problems in clinical research on the basis of only a small number of examples for which an expert decision is available. In digital pathology, large amounts of data are available, but providing expert knowledge for these data, e.g., in the form of markers in the data indicating the tissue type, as required for supervised learning approaches, is very time-consuming.
Both aspects are addressed in our research. Using the diagnosis of colorectal adenocarcinoma (colorectal cancer) as an application example, robust and adaptable AI methods for the automatic detection of tissue types (such as tumor tissue or muscle tissue) are being developed and researched in a specific manner.
Revolutionizing Digital Pathology with Few Data and Few Labels Learning Methods
The basis for the automatic segmentation of a tissue section into different tissue classes is a so-called "convolutional neural network" (CNN). This neural network is trained using sample images. To evaluate the robustness of various models for example, tissue sections were digitized with six different scanners. On these data the classification quality of the trained models is compared.
One approach to generate robust models is domain-specific data augmentation, as it is being further developed in the ADA Lovelace Center's Few Data Learning competence pillar. In data augmentation, additional images are artificially generated from existing reference images using specified transformations, such as changes in brightness or color tone. The basic idea here is to take into account the heterogeneity in the later application already during training by specifically manipulating the training images. Different augmentation techniques and their combinations are compared. The focus is on changes in the color values of the image. For example, the contrast and saturation of the images are changed, but application-specific color changes are also used.
Another focus is on the evaluation of so-called Few-Shot procedures from the Few Labels Learning competence pillar. In this method, the classes (in our case, tissue classes such as tumor) being distinguished can be subsequently adjusted based on a few annotated data without the need to retrain the neural network (competence pillar Few Label Learning). Different variants of so-called prototypical networks are used. The basic building block is a CNN as well. By this procedure now also new classes can be added subsequently. In each case, only a few examples (representatives) of the new class are needed and a class representation is calculated on the basis of these.
It turns out that a combination of the two methods is beneficial and increases the robustness of the prototypical networks on new data.